Diversity & Classification
Prokaryotic Family Tree
We want you to know that prokaryotes are diverse and we've got the data to back it up. A great way to demonstrate diversity is by graphing a phylogenetic tree. A phylogenetic tree is kind of like a family tree, but for different species of living things. At the root of the tree is a shared ancestor, and at the other end are all the relatives, both close and distant, of the current generation.If you want to see a good example of a family tree, we recommend checking out the British royal family tree. This family tree traces the links from a common ancestor (in this case, Queen Elizabeth II, who was born in 1926) through to her great grandchildren, including Prince George and Princess Charlotte.
The phylogenetic tree of life is similar to a family tree, except instead of going back to a single person, like Queen Elizabeth II, it's rooted in a shared ancestor for all of the species included. You may not look a lot like a bacterium, but you're very distantly related to it. Of course, any ancestor we shared with bacteria lived very, very, very long ago.
This is a diagram of the phylogenetic tree of life. It separates organisms based on their genomic sequences.

Phylogenetic Tree of Life. Image from here.
The root of the tree lies at the center bottom. This point represents the common ancestor of all living things. Modern organisms span the outer reaches of the tree, from Firmicutes (a type of bacteria) on the left to animals (like us!) on the right. This tree shows two important things:
- Prokaryotes are really diverse
- Archaea really are separate from bacteria and eukaryotes.
Looking at the tree, we see that archaea exist in a completely separate branch from bacteria. In fact, they're even grouped a little more closely with the eukaryotes than with the bacteria. This is because when it comes to the enzymes used for several of their most basic processes, notably the enzymes used for transcription and translation, archaea are more like eukaryotes than bacteria. Archaea and bacteria might look similar under a microscope (as you can see in some of the images below), but, genetically, they're made of different stuff.
In the first image below, bacterial cells appear next to a large plant cell. The archaea in the second image (which isn't at the same scale!) looks a lot like bacteria, but are a genetically distinct branch of life.
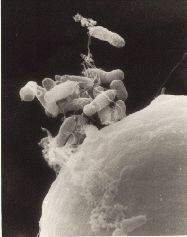
Image from here.

Image from here.
Prokaryotic Cells
So let's get into the differences between prokaryotic cells. Here's a simple schematic. These prokaryotes have two membranes, a cell wall between them, and, finally some DNA. It doesn't get much simpler than that. Actually, it does get a little simpler. Some prokaryotes don't even have the outer membrane (gasp!). We'll get into that in a bit.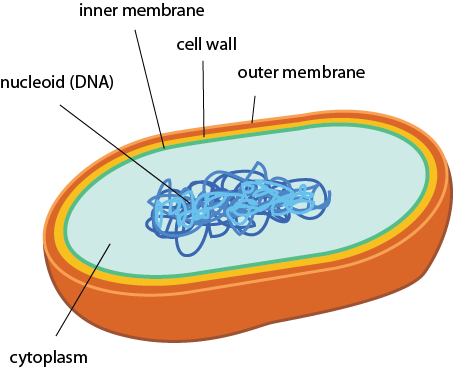
Simple, right?
Differences in the cell wall affect how prokaryotes grow and how we classify them. In archaea, the cell wall, called the S-layer (for surface layer), is made of proteins. In bacteria the cell wall is made up of a material called peptidoglycan. Peptidoglycan molecules consist of a peptide (peptides are like really small proteins) linked with a glycan (aka sugar).
Differences in prokaryotic cell walls also affect how other organisms respond to them. For example, the human immune system has specific receptors that recognize peptidoglycan, enabling it to detect bacteria (though sometimes not soon enough to keep us from feeling their effects). These receptors monitor the presence of bacteria in places where they should and shouldn't be. Think of them like tiny tracking devices that are keeping tabs on their comings and goings.
Gram Staining
Bacteria are divided into two major classes. This process is based on cell walls (are you sensing that these are important yet?). Hans Christian Gram, a Dane (who might have been great) worked in a German morgue and invented the staining process we use to detect which class a particular bacterium has. No relation to the sometimes-morbid Hans Christian Andersen, who wrote The Little Mermaid. Because the stain is named for a person, the first letter of Gram is always capitalized.The Gram stain is still used as the first step in identifying bacteria. The way it works is that a population of bacteria is exposed to a dye called crystal violet. Initially, this dye colors all the bacteria. When the dye is washed away, the Gram-positive bacteria, which have thick cell walls, hold all the dye in. Bacteria without those thick candy shells lose the dye and are classified as Gram negative. A weaker stain, called a counter stain, is then added so we can see the Gram negative bacteria in a pretty pink hue.
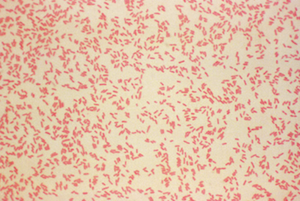
Gram-negative bacteria (rosy pink). Image from here.
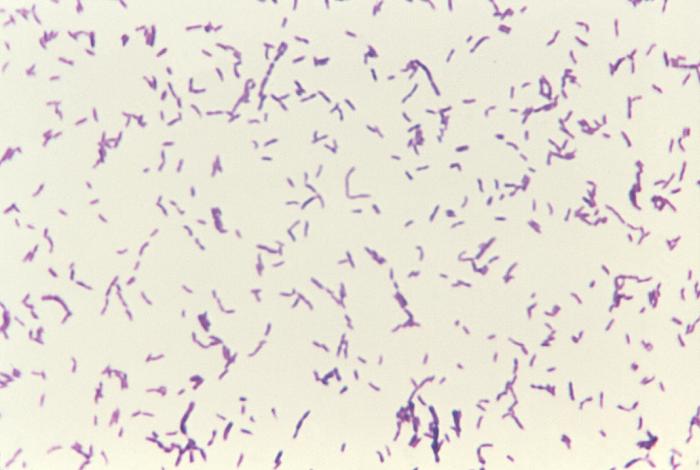
Gram-positive bacteria (royal purple). Image is ID# 3079 from the CDC Public Health Image Library.
Both Gram-negative and Gram-positive bacteria have similar cytoplasms and cytoplasmic membranes (also known as plasma membranes) made of phospholipid bilayers. Just like the ones eukaryotic and archaeal cells run around in. If you just can't get enough of plasma membranes, read up on 'em here.
Bottom line, the cell wall is where Gram negatives and Gram positives really show off their differences. Their different outsides strongly affect how they interact with the world. Even at the microbial level, appearances matter.
Gram-positive bacteria have a thick cell wall that surrounds their cytoplasmic membrane. In some species the wall is coated with polysaccharides in a final layer called the capsule. The capsule is slippery, helping bacteria slip and slide away from predators. It also blocks some viruses and detergents from harming the bacteria.
Gram-negative bacteria have an extra membrane, called the outer membrane, that surrounds their cell wall. The inner face of the outer membrane is composed of phospholipids, just like the inner membrane. The outer face is composed of a special material called lipopolysaccharide (LPS). Bet you can guess what LPS is made of too: it's part lipid and part polysaccharide (sugar). The polysaccharide part protrudes outside the cell. And just because they want to play slip and slide too, some Gram-negative bacteria have capsules.
The existence of an outer membrane means that Gram-negative bacteria have an extra cellular compartment between the two membranes. This region is called the periplasm, and it's where the bacterium hides the candy it doesn't want to share with its friends…wait…no. The periplasm is kind of like a giant cellular foyer or "airlock" that surrounds the cytoplasm, giving the cell an extra measure of control to what chemicals and nutrients are allowed to cross each of their membranes.
The Gram-negative bacterial cell wall is located in the periplasm. It is thinner than the Gram-positive cell wall. Gram-negative bacteria are okay with this because they get extra stability from the LPS in their outer membrane.

Gram-positive versus Gram-negative. Who will win?
Shapes
Prokaryotes, both bacteria and archaea, take on some inventive shapes, or morphologies. The shapes depend on the way their cell walls are constructed. The most common shapes are:- Rod-shaped (Bacillus)
- Spherical (Coccus)
- Spiral-shaped (Spirilla are rigid spirals; Spirochetes are flexible)
The images below are presented in false color in order to help distinguish the bacteria from the background (too bad these colors don't actually happen in nature). From top to bottom, they are Spheres, Rods, Spirals, and finally Curved Rods. We threw the curved rods (also called comma-shaped) in there just to point out that variations on the three major shapes are common.
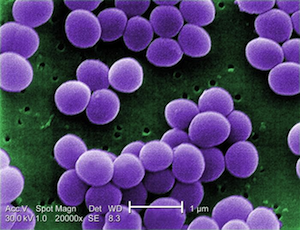
Cocci. Image from here.

Bacillus. Image from here.

Curved rod. Image from here.
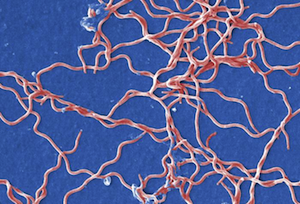
Spirochete. Image is ID# 13169 from the CDC Public Health Image Library.
DNA-based Prokaryotic Classification
A lot can be gained from understanding a bacterium's morphology. For example, if you think someone might have the disease cholera, but they don't have cells that look like the bacteria that causes cholera, you might want to try a different treatment. Or get a better doctor.Prokaryotes are a bit trickier to identify than multicellular eukaryotes. If you're trying to tell if a horse and a zebra are different organisms, all you need to do is open your eyes. We haven't seen too many stripy horses. Prokaryotes don't have stripes or as many obvious features to distinguish one from another, so scientists have figured out other ways of classifying them.
Serotyping
One way that prokaryotes are classified is by determining their serotypes. Serotype is determined by testing which antibodies (generally from an animal, like antibodies from a rabbit immune system) interact with a given sample. American microbiologist Rebecca Lancefield developed the serotype classification system in 1933.Different antibodies recognize different parts of molecules on the outsides of the prokaryotic cells. A major antigen responsible for serotyping in Gram-negative bacteria is the O-antigen, the polysaccharide component of LPS. While serotyping antibodies often recognize the outer regions of cells, they also respond to virulence factors (called exotoxins) that are released by pathogenic bacteria. Pathogenic bacteria are bacteria that can cause disease. We'll talk more about pathogens and exotoxins later.
Serotyping can be very sensitive and even identify different classes of bacteria of the same species, called strains. For example, the species Vibrio cholerae, which causes the disease cholera, has more than 200 serotypes. We pity the poor little bunny that figured that one out.
DNA Barcodes
Another method that is increasingly used is DNA sequencing. DNA sequencing can be used in one of two ways. In the first option, an entire bacterial genome sequence can be found and compared with known existing sequences. This is expensive, time consuming, and a bit boring if you're not into thousands of pages of As, Ts, Gs, and Cs. A quick and dirty approach involves only sequencing a specific region of the genome known to be variable across species. Such regions are sometimes called DNA barcodes, since they're unique to a given species. The DNA barcode sequence can be used like real-life barcodes to look up the organism attached to it.Brain Snack
DNA barcoding was used by high school students in New York to investigate if fish used for sushi were the actual fish advertised on the menu. In a big news story dubbed "Sushigate", the students, with the help of university researchers, found that 25% of fish were mislabeled. Check it out here.The work pioneered by these high school students continues. A more recent investigation in Los Angeles showed that over half of seafood there was labeled as some other fish.